In this example we want to download, import and plot over the 3500 km2 wide region of interest (mapped below) :
- a 1 year-long time series of monthly NDVI from MODIS Terra Vegetation indices (spatial resolution : 1 km ; temporal resolution : 1 month),
- the same 1 year-long times series of monthly precipitations from Global Precipitation Measurement (GPM) (spatial resolution : 1° ; temporal resolution : 1 month)
Check which collections are available
Before starting, let’s identify our collections of interest among the
collections that are available for download, using the function
mf_list_collections() :
collections <- mf_list_collections()
print(str(collections))
#> 'data.frame': 77 obs. of 26 variables:
#> $ collection : chr "GPM_3IMERGDE.06" "GPM_3IMERGDF.06" "GPM_3IMERGDF.07" "GPM_3IMERGDL.06" ...
#> $ source : chr "GPM" "GPM" "GPM" "GPM" ...
#> $ long_name : chr "GPM IMERG Early Precipitation L3 1 day 0.1 degree x 0.1 degree V06" "GPM IMERG Final Precipitation L3 1 day 0.1 degree x 0.1 degree V06" "GPM IMERG Final Precipitation L3 1 day 0.1 degree x 0.1 degree V07" "GPM IMERG Late Precipitation L3 1 day 0.1 degree x 0.1 degree V06" ...
#> $ nature : chr "Rainfall" "Rainfall" "Rainfall" "Rainfall" ...
#> $ provider : chr "NASA GES DISC" "NASA GES DISC" "NASA GES DISC" "NASA GES DISC" ...
#> $ url_metadata : chr "https://disc.gsfc.nasa.gov/datasets/GPM_3IMERGDE_06/summary" "https://disc.gsfc.nasa.gov/datasets/GPM_3IMERGDF_06/summary" "https://disc.gsfc.nasa.gov/datasets/GPM_3IMERGDF_07/summary" "https://disc.gsfc.nasa.gov/datasets/GPM_3IMERGDL_06/summary" ...
#> $ doi : chr "https://doi.org/10.5067/GPM/IMERGDE/DAY/06" "https://doi.org/10.5067/GPM/IMERGDF/DAY/06" "https://doi.org/10.5067/GPM/IMERGDF/DAY/07" "https://doi.org/10.5067/GPM/IMERGDL/DAY/06" ...
#> $ version : num 6.1 6.1 7 6.1 6.1 7 6.1 6.1 6.1 7 ...
#> $ spatial_resolution_m : int 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 ...
#> $ temporal_resolution : int 1 1 1 1 30 30 30 30 1 1 ...
#> $ temporal_resolution_unit: chr "day" "day" "day" "day" ...
#> $ spatial_coverage : chr "Global" "Global" "Global" "Global" ...
#> $ start_date : chr "2000-06-01" "2000-06-01" "2000-06-01" "2000-06-01" ...
#> $ end_date : chr "ongoing" "ongoing" "ongoing" "ongoing" ...
#> $ indicative_latency_days : int 1 100 100 1 100 100 0 1 100 100 ...
#> $ url_manual_access : chr "https://search.earthdata.nasa.gov/search?q=GPM_3IMERGDE_06" "https://search.earthdata.nasa.gov/search?q=GPM_3IMERGDF_06" "https://search.earthdata.nasa.gov/search?q=GPM_3IMERGDF_07" "https://search.earthdata.nasa.gov/search?q=GPM_3IMERGDL_06" ...
#> $ status : chr "Implemented" "Implemented" "Implemented" "Implemented" ...
#> $ login : chr "earthdata" "earthdata" "earthdata" "earthdata" ...
#> $ url_opendapserver : chr "https://gpm1.gesdisc.eosdis.nasa.gov/opendap/GPM_L3/" "https://gpm1.gesdisc.eosdis.nasa.gov/opendap/GPM_L3/" "https://gpm1.gesdisc.eosdis.nasa.gov/opendap/GPM_L3/" "https://gpm1.gesdisc.eosdis.nasa.gov/opendap/GPM_L3/" ...
#> $ url_programmatic_access : chr "https://gpm1.gesdisc.eosdis.nasa.gov/opendap/GPM_L3/GPM_3IMERGDE.06/" "https://gpm1.gesdisc.eosdis.nasa.gov/opendap/GPM_L3/GPM_3IMERGDF.06/" "https://gpm1.gesdisc.eosdis.nasa.gov/opendap/GPM_L3/GPM_3IMERGDF.07/" "https://gpm1.gesdisc.eosdis.nasa.gov/opendap/GPM_L3/GPM_3IMERGDL.06/" ...
#> $ url_opendapexample : chr "https://gpm1.gesdisc.eosdis.nasa.gov/opendap/GPM_L3/GPM_3IMERGDE.06/2000/06/3B-DAY-E.MS.MRG.3IMERG.20000601-S00"| __truncated__ "https://gpm1.gesdisc.eosdis.nasa.gov/opendap/hyrax/GPM_L3/GPM_3IMERGDF.06/2000/06/3B-DAY.MS.MRG.3IMERG.20000601"| __truncated__ "https://gpm1.gesdisc.eosdis.nasa.gov/opendap/hyrax/GPM_L3/GPM_3IMERGDF.07/2000/06/3B-DAY.MS.MRG.3IMERG.20000601"| __truncated__ "https://gpm1.gesdisc.eosdis.nasa.gov/opendap/hyrax/GPM_L3/GPM_3IMERGDL.06/2000/06/3B-DAY-L.MS.MRG.3IMERG.200006"| __truncated__ ...
#> $ dim_lon : chr "lon" "lon" "lon" "lon" ...
#> $ dim_lat : chr "lat" "lat" "lat" "lat" ...
#> $ dim_time : chr "time" "time" "time" "time" ...
#> $ dim_proj : chr NA NA NA NA ...
#> $ crs : chr "+proj=longlat +datum=WGS84 +no_defs +type=crs" "+proj=longlat +datum=WGS84 +no_defs +type=crs" "+proj=longlat +datum=WGS84 +no_defs +type=crs" "+proj=longlat +datum=WGS84 +no_defs +type=crs" ...
#> NULLSo our collections of interest are :
- MOD13A3.061 : MODIS/Terra Vegetation Indices Monthly L3 Global 1 km SIN Grid
- GPM_3IMERGM.07 : GPM IMERG Final Precipitation L3 1 month 0.1 degree x 0.1 degree V07
Setup the region and time range of interest
First we prepare the script : define the ROI, the time frame, and the folder were the data will be downloaded :
# Set ROI and time range of interest
roi <- st_as_sf(data.frame(id="Korhogo", geom="POLYGON ((-5.82 9.54, -5.42 9.55, -5.41 8.84, -5.81 8.84, -5.82 9.54))"), wkt="geom", crs = 4326)
time_range <- as.Date(c("2017-01-01","2017-12-31"))Login to EOSDIS Earthdata
And we login to EOSDIS Earthdata with our credentials. To create an account go to : https://urs.earthdata.nasa.gov/.
Get the URLs of the data to download
With the function mf_get_url(), we get the https URLs
for our collections of interest (MOD11A1.061 and
GPM_3IMERGDF.07) given our ROI and time frame.
:arrow_right: For each collection, we can select a subset only of
variables (sometimes called “dimensions”, or “bands”) that are of
interest for us, through the parameter variables. By
default, this parameter is set to NULL, which means that
all the available variables for the specified collection are included.
To get the variables available for a given collection along with
information for each of them (description, etc.), use the function
mf_list_variables().
For more information on the variables in the context of OPeNDAP, see the OPeNDAP Data model documentation.
## Get the URLs of MODIS Vegetation indexes
urls_mod11a1 <- mf_get_url(
collection = "MOD13A3.061",
variables = c("_1_km_monthly_NDVI","_1_km_monthly_EVI","_1_km_monthly_VI_Quality"), # get the available variables with : mf_list_variables("MOD13A3.061"), or set to NULL (default) to download all the variables
roi = roi,
time_range = time_range )
#> Building the URLs...
#> Estimated maximum size of data to be downloaded is 1 Mb
## Get the URLs of GPM daily
urls_gpm <- mf_get_url(
collection = "GPM_3IMERGM.07",
variables = c("precipitation"), # get the available variables with : mf_list_variables("GPM_3IMERGM.07")
roi = roi,
time_range = time_range )
#> Building the URLs...
#> Estimated maximum size of data to be downloaded is 1 Mb
nrow(urls_mod11a1)
#> [1] 1
urls_mod11a1
#> id_roi time_start collection name
#> 1 Korhogo 2017-01-01 MOD13A3.061 MOD13A3.061.2017001_2017335.h17v08.nc4
#> url
#> 1 https://opendap.cr.usgs.gov/opendap/hyrax/MOD13A3.061/h17v08.ncml.nc4?MOD_Grid_monthly_1km_VI_eos_cf_projection,_1_km_monthly_NDVI%5B203:214%5D%5B55:140%5D%5B511:561%5D,_1_km_monthly_EVI%5B203:214%5D%5B55:140%5D%5B511:561%5D,_1_km_monthly_VI_Quality%5B203:214%5D%5B55:140%5D%5B511:561%5D,time%5B203:214%5D,YDim%5B55:140%5D,XDim%5B511:561%5D
#> maxFileSizeEstimated
#> 1 561000
nrow(urls_gpm)
#> [1] 12
head(urls_gpm,3)
#> id_roi time_start collection
#> 1 Korhogo 2017-01-01 GPM_3IMERGM.07
#> 2 Korhogo 2017-02-01 GPM_3IMERGM.07
#> 3 Korhogo 2017-03-01 GPM_3IMERGM.07
#> name
#> 1 3B-MO.MS.MRG.3IMERG.20170101-S000000-E235959.01.V07B.nc4
#> 2 3B-MO.MS.MRG.3IMERG.20170201-S000000-E235959.02.V07B.nc4
#> 3 3B-MO.MS.MRG.3IMERG.20170301-S000000-E235959.03.V07B.nc4
#> url
#> 1 https://gpm1.gesdisc.eosdis.nasa.gov/opendap/GPM_L3/GPM_3IMERGM.07/2017/3B-MO.MS.MRG.3IMERG.20170101-S000000-E235959.01.V07B.HDF5.nc4?precipitation%5B0:0%5D%5B1742:1746%5D%5B989:996%5D,time%5B0:0%5D,lon%5B1742:1746%5D,lat%5B989:996%5D
#> 2 https://gpm1.gesdisc.eosdis.nasa.gov/opendap/GPM_L3/GPM_3IMERGM.07/2017/3B-MO.MS.MRG.3IMERG.20170201-S000000-E235959.02.V07B.HDF5.nc4?precipitation%5B0:0%5D%5B1742:1746%5D%5B989:996%5D,time%5B0:0%5D,lon%5B1742:1746%5D,lat%5B989:996%5D
#> 3 https://gpm1.gesdisc.eosdis.nasa.gov/opendap/GPM_L3/GPM_3IMERGM.07/2017/3B-MO.MS.MRG.3IMERG.20170301-S000000-E235959.03.V07B.HDF5.nc4?precipitation%5B0:0%5D%5B1742:1746%5D%5B989:996%5D,time%5B0:0%5D,lon%5B1742:1746%5D,lat%5B989:996%5D
#> maxFileSizeEstimated
#> 1 112
#> 2 112
#> 3 112Download the data
Now we download the data with the function
mf_download_data().
The destination file for each dataset is specified in the column destfile of the dataframes urls_mod11a1 and urls_gpm. A destination file is specified by default but it can of course be modified.
Setting the argument parallel to TRUE will
parallelize - therefore fasten - the download in case numerous datasets
need to be downloaded.
# Hint : to fasten the download in case of multiple collections, bind them in a single data.frame (as shown below) before executing the function 'mf_download_data()' :
df_to_dl <- rbind(urls_mod11a1, urls_gpm)
# Then download the data (by default, in temporary directory, but the target folder can be set with the argument 'path'
res_dl <- mf_download_data(df_to_dl = df_to_dl,
parallel = TRUE)
#> 13 datasets in total : 0 already downloaded and 13 datasets to download
#> Downloading the data ... (destination folder: /tmp/Rtmp2vpw2a/modisfast_3df01416cf7b )
#> Estimated maximum size of data to be downloaded is ~ 1 Mb
#>
#> Actual size of downloaded data is 1 Mb
print(str(res_dl))
#> 'data.frame': 13 obs. of 10 variables:
#> $ id_roi : chr "Korhogo" "Korhogo" "Korhogo" "Korhogo" ...
#> $ time_start : Date, format: "2017-01-01" "2017-01-01" ...
#> $ collection : chr "MOD13A3.061" "GPM_3IMERGM.07" "GPM_3IMERGM.07" "GPM_3IMERGM.07" ...
#> $ name : chr "MOD13A3.061.2017001_2017335.h17v08.nc4" "3B-MO.MS.MRG.3IMERG.20170101-S000000-E235959.01.V07B.nc4" "3B-MO.MS.MRG.3IMERG.20170201-S000000-E235959.02.V07B.nc4" "3B-MO.MS.MRG.3IMERG.20170301-S000000-E235959.03.V07B.nc4" ...
#> $ url : chr "https://opendap.cr.usgs.gov/opendap/hyrax/MOD13A3.061/h17v08.ncml.nc4?MOD_Grid_monthly_1km_VI_eos_cf_projection"| __truncated__ "https://gpm1.gesdisc.eosdis.nasa.gov/opendap/GPM_L3/GPM_3IMERGM.07/2017/3B-MO.MS.MRG.3IMERG.20170101-S000000-E2"| __truncated__ "https://gpm1.gesdisc.eosdis.nasa.gov/opendap/GPM_L3/GPM_3IMERGM.07/2017/3B-MO.MS.MRG.3IMERG.20170201-S000000-E2"| __truncated__ "https://gpm1.gesdisc.eosdis.nasa.gov/opendap/GPM_L3/GPM_3IMERGM.07/2017/3B-MO.MS.MRG.3IMERG.20170301-S000000-E2"| __truncated__ ...
#> $ maxFileSizeEstimated: num 561000 112 112 112 112 112 112 112 112 112 ...
#> $ destfile : chr "/tmp/Rtmp2vpw2a/modisfast_3df01416cf7b/data/Korhogo/MOD13A3.061/MOD13A3.061.2017001_2017335.h17v08.nc4" "/tmp/Rtmp2vpw2a/modisfast_3df01416cf7b/data/Korhogo/GPM_3IMERGM.07/3B-MO.MS.MRG.3IMERG.20170101-S000000-E235959.01.V07B.nc4" "/tmp/Rtmp2vpw2a/modisfast_3df01416cf7b/data/Korhogo/GPM_3IMERGM.07/3B-MO.MS.MRG.3IMERG.20170201-S000000-E235959.02.V07B.nc4" "/tmp/Rtmp2vpw2a/modisfast_3df01416cf7b/data/Korhogo/GPM_3IMERGM.07/3B-MO.MS.MRG.3IMERG.20170301-S000000-E235959.03.V07B.nc4" ...
#> $ fileDl : logi TRUE TRUE TRUE TRUE TRUE TRUE ...
#> $ fileSize : num 288607 26540 26495 26548 26560 ...
#> $ dlStatus : num 1 1 1 1 1 1 1 1 1 1 ...
#> NULLImport the data in R
Various packages and related classes can be used to read the data
downloaded through OPeNDAP. If terra
(successor of raster)
is surely the most famous class for raster objects, many packages
facilitate the use of spatiotemporal data cubes in formats such as those
proposed through modisfast (e.g. NetCDF). For instance,
MODIS or VIIRS products can be imported as a stars object
from the excellent stars
package for data cubes manipulation. All the data can also be imported
as ncdf4 objects using e.g. the ncdf4
package.
:warning: :warning: :warning: :warning:
Important warning regarding the use of the data after download
through modisfast
Care must be taken when importing data that was downloaded through
the OPeNDAP data providers servers. Depending on the collection, some
“issues” were raised. These issues are independent from
modisfast : they result most of time of a kind of lack of
full implementation of the OPeNDAP framework by the data providers.
These issues are :
- for MODIS and VIIRS collections : CRS has to be provided
- for GPM collections : CRS has to be provided + data have to be flipped
End warning
These issues can easily be dealt at the import phase in R using the
function mf_import_data(). This function enables to open
the data in R as a SpatRaster object (from the package terra)
:
# import MOD11A1.061
# argument 'path' is the folder where the data are stored (here, in a temporary directory)
modis_ts <- mf_import_data(path = dirname(list.files(path = tempdir(), pattern = "MOD13A3.061", recursive = TRUE, full.names = TRUE)),
collection = "MOD13A3.061",
proj_epsg = 4326)
#> Importing the dataset as a SpatRaster object...
# import GPM
gpm_ts <- mf_import_data(path = unique(dirname(list.files(path = tempdir(), pattern = "3IMERG", recursive = TRUE, full.names = TRUE))),
collection = "GPM_3IMERGM.07",
proj_epsg = 4326)
#> Importing the dataset as a SpatRaster object...
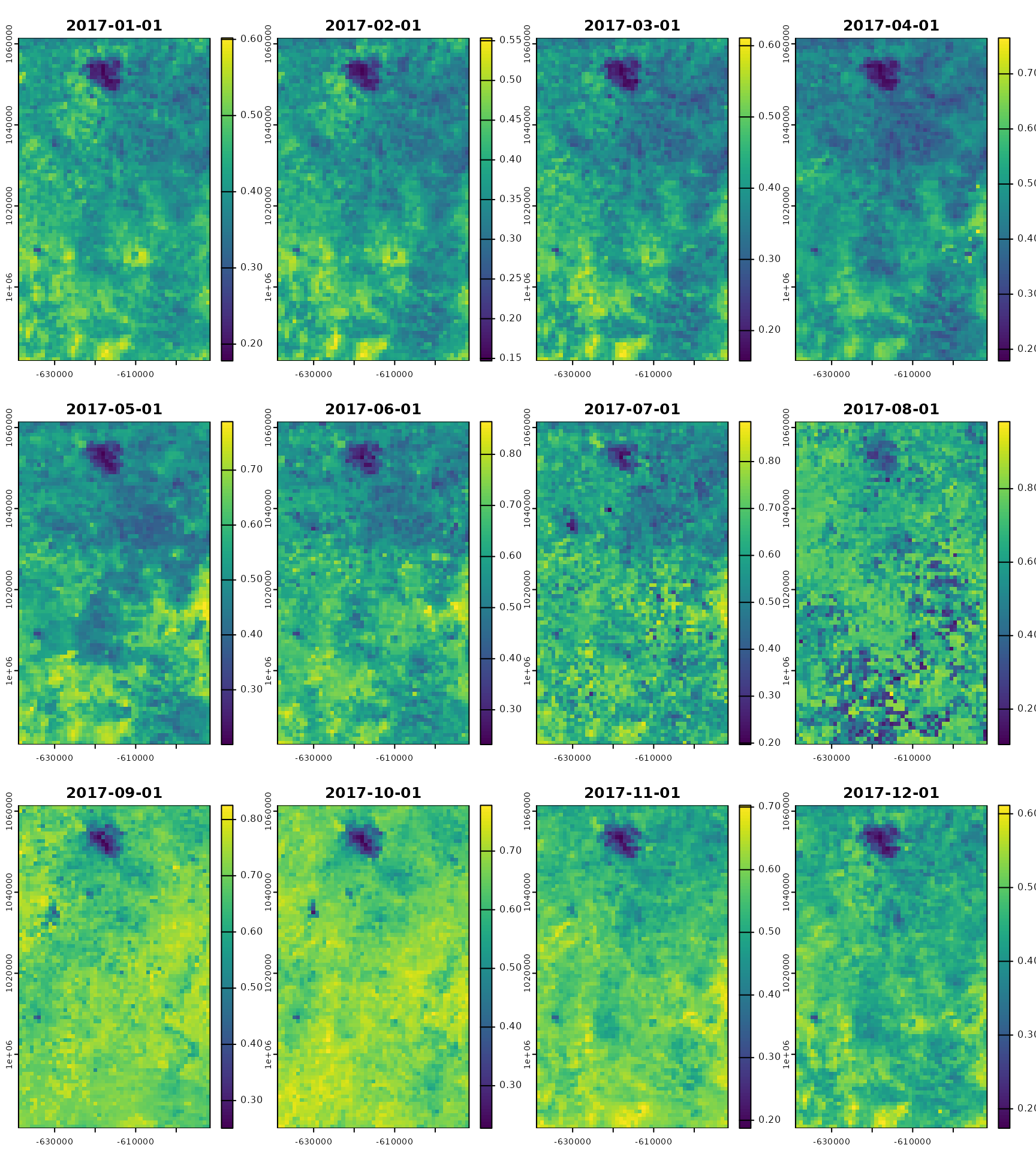
modis_ts
#> class : SpatRaster
#> dimensions : 85, 52, 36 (nrow, ncol, nlyr)
#> resolution : 0.008420659, 0.008420659 (x, y)
#> extent : -5.826512, -5.388637, 8.830077, 9.545833 (xmin, xmax, ymin, ymax)
#> coord. ref. : lon/lat WGS 84 (EPSG:4326)
#> source(s) : memory
#> names : _1_km~EVI_1, _1_km~EVI_2, _1_km~EVI_3, _1_km~EVI_4, _1_km~EVI_5, _1_km~EVI_6, ...
#> min values : 0.1065465, 0.09200972, 0.09880671, 0.1148083, 0.1415658, 0.1807865, ...
#> max values : 0.3287283, 0.33806026, 0.36024132, 0.4717904, 0.5491539, 0.5933855, ...
#> unit : EVI, EVI, EVI, EVI, EVI, EVI, ...
#> time (days) : 2017-01-01 to 2017-12-01
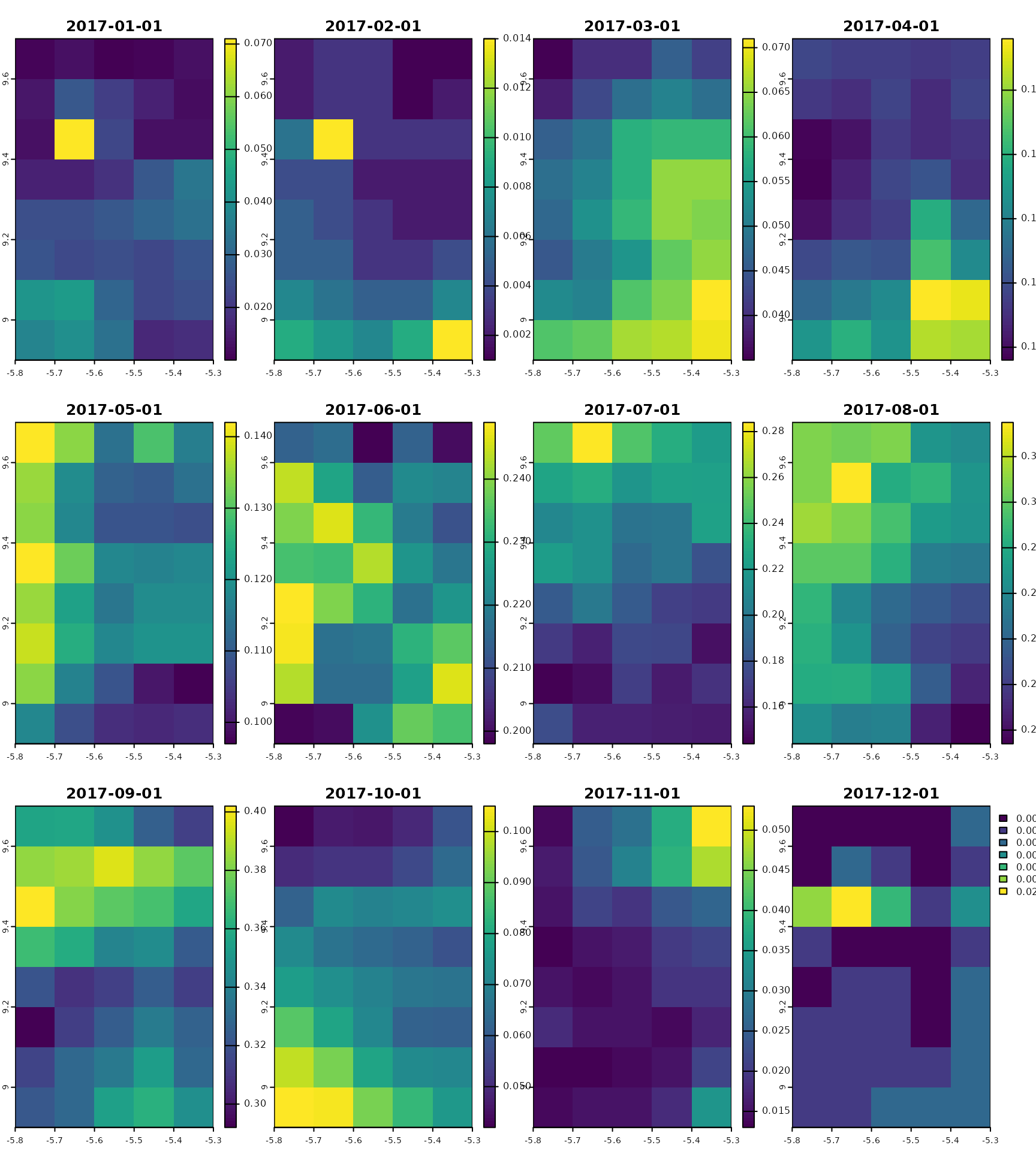
gpm_ts
#> class : SpatRaster
#> dimensions : 8, 5, 12 (nrow, ncol, nlyr)
#> resolution : 0.09999999, 0.09999999 (x, y)
#> extent : -5.8, -5.3, 8.9, 9.7 (xmin, xmax, ymin, ymax)
#> coord. ref. : lon/lat WGS 84 (EPSG:4326)
#> source(s) : memory
#> names : preci~ation, preci~ation, preci~ation, preci~ation, preci~ation, preci~ation, ...
#> min values : 0.01000000, 0.00100000, 0.03500000, 0.0960000, 0.09700001, 0.198, ...
#> max values : 0.07099995, 0.01399999, 0.07099999, 0.1959999, 0.14200000, 0.249, ...
#> time : 2017-01-01 to 2017-12-01 UTC